Up : MAXI Data Analysis
Previous : Data Analysis
Tips, Tricks, and Traps
This page describes what's happening when MAXI ftools are running, and what users should be careful to obtain confident results.
Contents
1. What's mxproduct?
mxproduct is a perl script running the MAXI ftools to generate images, light curves, spectra, and response files, calling the general FTOOLS and the following new MAXI ftools :
- mxextract ... Create a single standard event file from plural region event files for a given coordinates (RA, DEC) and radius.
- mxscancur ... Create an observation and instrument specific file to describe observational conditions.
- mxgscandat, mxsscandat ... Create observation and instrument specific scan history files for GSC and SSC.
- mxlcscan ... Create light curve files.
- mxgtiwmap ... Create an exposure histogram for each incident angle of a given input target.
- mxgrmfgen, mxsrmfgen ... Create a response file for GSC and SSC.
EXAMPLE
% mxdownload_wget -coordinates 83.633083,22.0145 -date_from 2010-02-01 -date_to 2010-02-05 (downloads data)
% mxproduct object=crab 83.633083 22.0145 2010-02-01 2010-02-05
> mxextract 'inst=gsc' 'infname=@crab_g_low_evt.list' 'split=1' 'outfname=crab' 'outfstem=crab' 'object=crab' 'ra=83.633083' 'dec=22.014500' \
'radi_i=0' 'radi_o=8' 'nx=200' 'ny=200' 'pixsize=0.1' 'leapsecfile=REFDATA' 'attlist=trend/attlist.fits'
> mxscancur 'instr=GSC' 'outfname=products/crab_g_low_scancur.fits' 'ra=83.633083' 'dec=22.014500' 'tstart=55197' 'tstop=55199' 'dt=1' \
'coltha_max=5.0' 'attlist=trend/attlist.fits' 'obscol_dname=/heasoft/6.25/x86_64-pc-linux-gnu-libc2.12/refdata' \
'sarjfnamelist=trend/isalist.fits' 'pmfnamelist=trend/isplist.fits' 'gtiskip=0' \
'gtifile0=trend/stdgti/gsc_low/mx_gsc0.gti.gz' 'gtifile1=trend/stdgti/gsc_low/mx_gsc1.gti.gz' \
'gtifile2=trend/stdgti/gsc_low/mx_gsc2.gti.gz' 'gtifile3=trend/stdgti/gsc_low/mx_gsc3.gti.gz' \
'gtifile4=trend/stdgti/gsc_low/mx_gsc4.gti.gz' 'gtifile5=trend/stdgti/gsc_low/mx_gsc5.gti.gz' \
'gtifile6=trend/stdgti/gsc_low/mx_gsc6.gti.gz' 'gtifile7=trend/stdgti/gsc_low/mx_gsc7.gti.gz' \
'gtifile8=trend/stdgti/gsc_low/mx_gsc8.gti.gz' 'gtifile9=trend/stdgti/gsc_low/mx_gsc9.gti.gz' \
'gtifile10=trend/stdgti/gsc_low/mx_gsca.gti.gz' 'gtifile11=trend/stdgti/gsc_low/mx_gscb.gti.gz' \
'leapsecfile=REFDATA' 'ssdockinfo=CALDB' 'teldefgsc=CALDB' 'teldefssc=CALDB' 'coleagsc=CALDB' \
'coleassc=CALDB' 'clobber=YES'
> mxgscandat 'scanflist=products/crab_g_low_scancur.fits' 'outfname=crab_g_low_scandat.fits' 'pibandfname=crab_g_low_pi_ch.list' \
'evtflist=products/crab_g_low.evt' 'bevtflist=products/crab_g_low.evt' 'srcregfile=NONE' 'bgdregfile=NONE' 'bgdscale=-1' 'mjd_start=55197' 'mjd_end=55199' \
'phindx=2.1' 'nh=0.35' 'src_dcolphi_max=1.7' 'src_dcoltha_max=1.5' 'bgd_dcoltha_min=1.5' 'bgd_dcoltha_max=4.5' 'evt_colphi_max=38.2' 'dlpath=0' \
'effcalc=1' 'gtifile0=NONE' 'gtifile1=products/crab_g1_low_gti.fits' 'gtifile2=products/crab_g2_low_gti.fits' 'gtifile3=NONE' 'gtifile4=NONE' \
'gtifile5=NONE' 'gtifile6=NONE' 'gtifile7=products/crab_g7_low_gti.fits' 'gtifile8=products/crab_g8_low_gti.fits' 'gtifile9=NONE' 'gtifile10=NONE' 'gtifile11=NONE'\
'leapsecfile=REFDATA' 'ldparm=CALDB' 'hvbithist=CALDB' 'mfptabbe=CALDB' 'mfptabxe=CALDB' 'ssdockinfo=CALDB' 'clobber=YES'
> mxlcscan 'scandatfname=products/crab_g_low_scandat.fits' 'outfname=crab_g_low.lc' 'abs_colphi_min=1.0' 'abs_colphi_max=36.2' \
'srctime_min=1.0' 'bgdtime_min=1.0' 'colexp_min=20.0' 'scnts_min=1' 'bcnts_min=1' 'camlist=1278' 'hvsel=0' 'moondist=8' 'clobber=YES'
> mxgtiwmap 'scanfname=products/crab_g_low_scancur.fits' 'outfname=crab_g_low_wmap.fits' 'mxgtifname=products/crab_g_low_gti.fits' \
'mjd_start=55197' 'mjd_end=55199' 'leapsecfile=REFDATA' 'colssc=CALDB'
> mxgrmfgen 'wmapfname=products/crab_g_low_wmap.fits' 'specfname=products/crab_g_low_src.pi' 'bgdfname=products/crab_g_low_bgd.pi' 'rmfname=crab_g_low.rsp'\
'gsclist=1278' 'dlpath=0' 'hvsel=0' 'leapsecfile=REFDATA' 'arfcorr=CALDB' 'hvbithist=CALDB' 'rmffile=CALDB' 'clobber=YES'
2. What's mxproduct doing?
Outline Flowchart of
mxproduct with default settings (GSC).
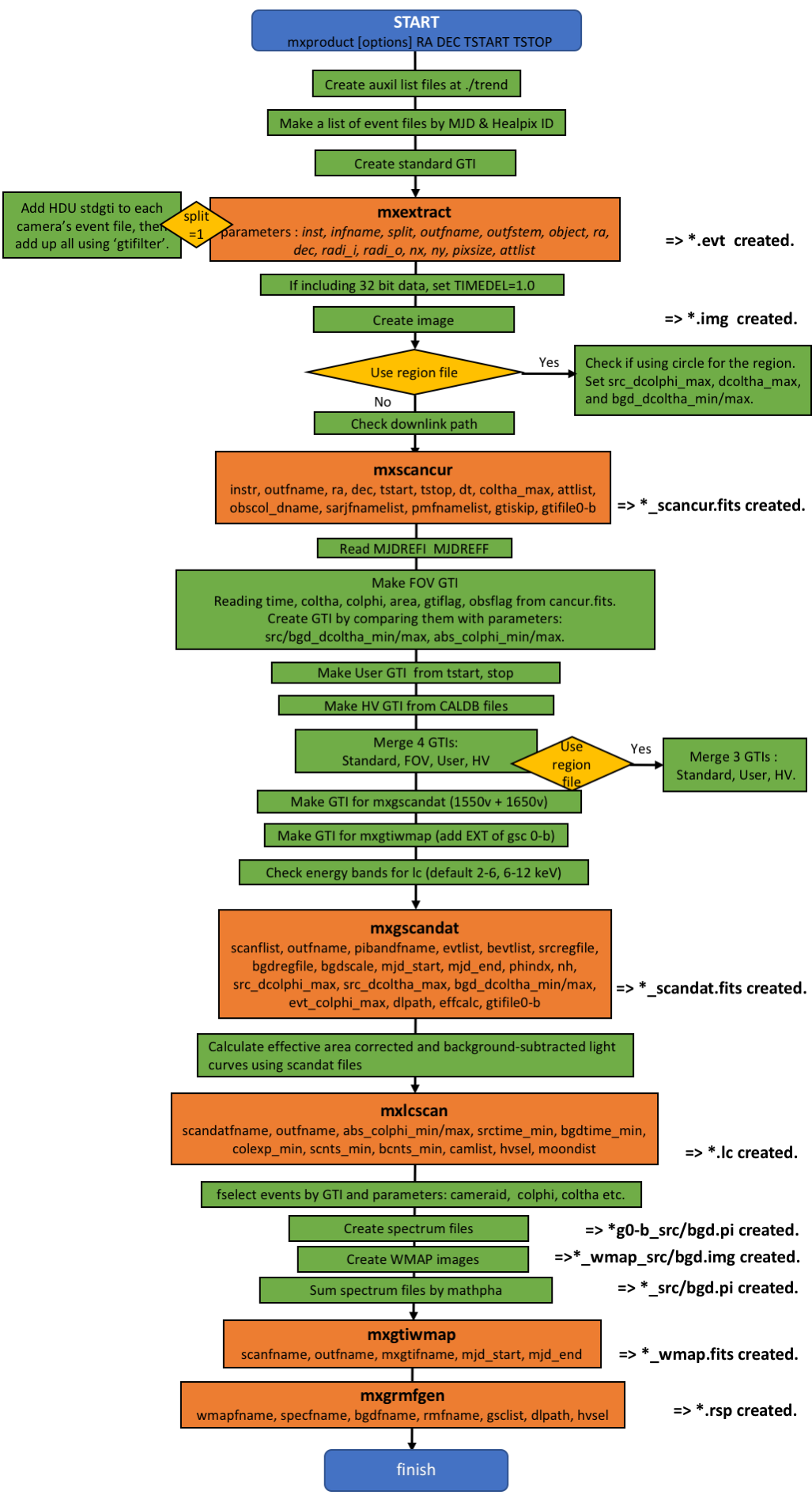
3. What mxproduct has created during the process?
mxproduct creates science products (see the
Data Analysis page for details).
Output files for GSC with object=crab
products/- crab_g_low.evt
- crab_g_low.img
- crab_g_low_wmap_src.img
- crab_g_low_wmap_bgd.img
- crab_g_low_src.pi
- crab_g_low_bgd.pi
- crab_g_low.rsp
- crab_g_low.lc
- crab_g_low_scancur.fits
- mxproduct.log
|
Output files for SSC with object=crab skip_ssc=0(default:1, skip)
products/- crab_s_med.evt
- crab_s_med.img
- crab_s_med_wmap_bgd.img
- crab_s_med_wmap_src.img
- crab_s_med_src.pi
- crab_s_med_bgd.pi
- crab_s_med.rsp
- crab_s_med.lc
- crab_s_med_scancur.fits
- mxproduct.log
|
mxproduct also creates the following intermediate files for the use of the processings and calculations, which are deleted at the end of the process. Users can leave them by running
mxproduct with an option, cleanup=no.
Intermediate files for GSC with cleanup=no(default)
at the directory you ran mxproduct
- crab_g_low_evt.list
- crab_g_low_camera.list
- crab_g_low_pi_ch.list
products/- crab_g_low_scandat.fits
- crab_g_low_wmap.fits
- crab_g[0-b]_low_[1550:1650]v_[src:bgd].pi
- crab_g[0-b]_low_[1550:1650]v_[src:bgd].evt
- crab_g[0-b]_low.evt
- crab_g_low_gti.fits
- crab_user_gti.fits
- crab_g[0-b]_low_[1550:1650]v_hvgti.fits
- crab_g[0-b]_low_fovgti.fits
- crab_g[0-b]_low_fovgti_[src:bgd].fits
- crab_g[0-b]_low_[1550:1650]v_gti.fits
- crab_g[0-b]_low_[1550:1650]v_gti_[src:bgd].fits
- crab_g[0-b]_low_gti.fits
- crab_g[0-b]_low_gti_[src:bgd].fits
|
Intermediate files for SSC with -no-cleanup=1(default:0)
at the directory you ran mxproduct
- crab_s_med_evt.list
- crab_s[h:z]_med.list
- crab_s_med_pi_ch.list
products/- crab_s_med_scandat.fits
- crab_s_med_wmap.fits
- crab_s[h:z]_med_[src:bgd].pi
- crab_s[h:z]_med_[src:bgd].evt
- crab_s[h:z]_med.evt
- crab_s[h:z]_med.rsp (SSC only)
- crab_s[h:z]_med.img (SSC only)
- crab_s_med_gti.fits
- crab_user_gti.fits
- crab_s[h:z]_med_fovgti_[src:bgd].fits
- crab_s[h:z]_med_gti.fits
- crab_s[h:z]_med_gti_[src:bgd].fits
|
4. What's written in command.log?
Commands which mxproduct is actually running, and comments on what mxproduct is doing. Users can refer the commands, and re-create science files as you like. For particular analysis, which users sometimes want to do, we will summarize how-tos in the Thread page.
5. Caveats
- · mxdownload_wget download the data including a day after the specified STOP date (because of the requirement of mxproduct).
- · The GSC low (gsc_low/) data-set is the default. GSC med (gsc_med) is also available for most of the time, but not calibrated up to 30 keV.
- · The generated images are neither exposure-corrected nor background-subtracted.
- · The source and background region files allow only circles. Other shapes are not accepted. If the background region is far from the source, the background may be estimated wrong.
- · When you exclude neighbouring sources from the source/background region, the effective area may not be calculated correctly.
- · The generated light-curves are not corrected for the effective area. The absolute count rate, especially in the low energy band, may be underestimated up to ~10%. The light-curves may show occasional artificial jumps.
- · MAXI does not cover all the 768 sky regions (HEALPix) everyday; some regions may not be available on a particular day. Also, there is a lack of data for different reasons: MAXI shutdown, downlink problems, processing errors etc. The detailed information about those problems will be appeared.
- · mxproduct calculates exposure at the target position, RA and DEC specified in the command line.
It is assumed that the region file to extract the events, if specified, is centered and covers the entire PSF.
When the center position of the region file is shifted from the center, and/or part of the PSF are missing (e.g. to exclude a contaminating source), the exposure may not be correct.
Even in such a case, the background scale factor is correct, and the background subtracted spectra and light-curves are calculated.
- · If a neighbouring object becomes X-ray brightening unexpectedly, the background flux would be higher, and the target flux would be estimated apparently lower.
- · A realistic simulation under optimal observation conditions suggests that MAXI will provide all-sky images of X-ray sources of ~20 mCrab ( 7e-10 erg/cm2/s in the energy band of 2?30 keV) from observations during one ISS orbit (90 min), ~4.5 mCrab for one day, and ~2 mCrab for one week. (Matsuoka et al. PASJ 61, 999, 2009)
- · mxproduct runs without region files, however, the estimated background level might be underestimated. We recommend using options, --srcregfile-gsc, --bgdregfile-gsc, --srcregfile-ssc, --bgdregfile-ssc to specify regions.

